nano-Differential Scanning Fluorimetry (nanoDSF)
nano-Differential scanning fluorimetry, or nanoDSF, is a biophysical characterization technique used for assessing the conformational stability of a biological sample.
It uses the intrinsic fluorescence of a protein to monitor how it responds to stress inputs such as temperature or chaotropes. This information is used to determine conformational stability of the protein, and to rank candidates or buffer formulations based on their impact on this stability.
nanoDSF is a valuable tool for many researchers in therapeutic spaces
nanoDSF is a useful tool for anyone developing biologics or gene therapies. Complex biological samples such as enzymes, monoclonal antibodies, or AAVs that will be used for treatments must be highly stable for storage, transport, and clinical administration.
nanoDSF enables you to monitor the conformational stability of your sample. This information helps you optimize your sample by making changes to the sequence or buffer environment, and measuring how those changes impact stability. Stability parameters are used to rank candidates for therapies and select those with greater stability for further characterization and development.
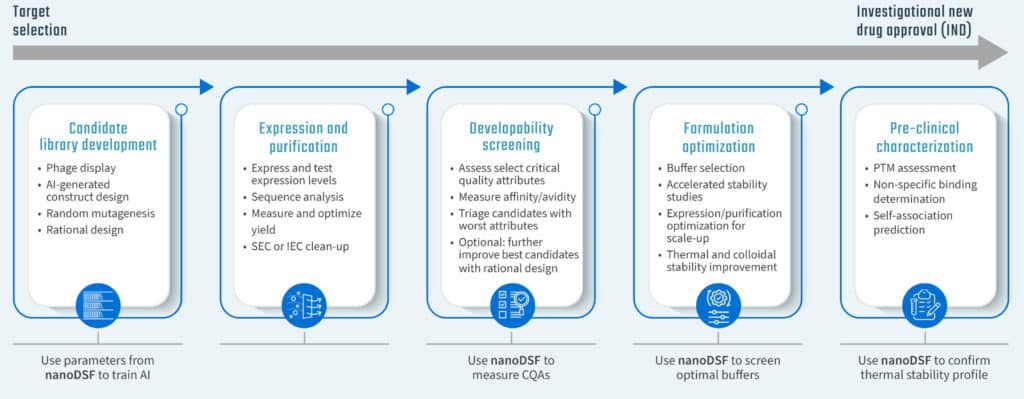
Apply nanoDSF measurements in many aspects of therapeutic development, including:
- During expression and purification
- Pre-formulation phase
- Developability assessment
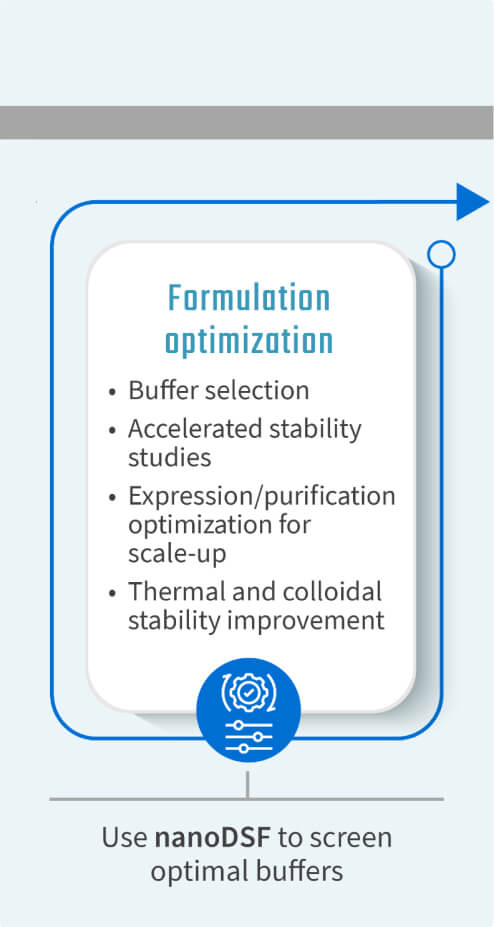
- Buffer formulation and optimization
- Comparability studies during scale-up or process changes




nanoDSF provides insight about the stability of your samples
Conformational stability indicates how likely a protein sample is to remain folded, and how well it holds up in stressful conditions. When a thermal or chemical denaturant gradient is applied to a protein sample, it will begin to unfold under thermal or chemical duress. Researchers use the point at which unfolding occurs as an indication of the overall impact a modification or condition has on stability.
Changes to the primary sequence or buffer environment impact the stability of a protein. When screening libraries of candidates or conditions, it’s important to only pass on candidates with improved stability. The parameters generated from nanoDSF data enable ranking candidates for increased stability attributes.
Melting temperature, Tm
Melting temperature, or Tm, is the temperature along a thermal denaturation gradient at which 50% of the protein is folded, and 50% is unfolded. Tm is a commonly used parameter for ranking the most thermostable candidates – the higher the Tm, the more stable the candidate, and the more optimal it is for further development.
Onset of unfolding, Ton
Onset of unfolding, or Ton, is the temperature at which a protein begins to unfold. Modifications to the protein sequence or buffer do not always impact the Tm. Ton is an additional parameter to indicate the change in thermal stability when changes are made to a candidate.
Slope at IP (inflection point)

Energy of unfolding, ΔG, is an energetic value parameter that tells you how much energy input is required to trigger unfolding of your sample at a given temperature. Protein unfolding for most proteins used in labs is not energetically favorable, so energy is required to disrupt the folding; the more positive ΔG, the more energy is required to trigger unfolding. Change in ΔG, ΔΔG is used to compare the change in ΔG between two different conditions.
nanoDSF generates information about your sample using its intrinsic fluorescence
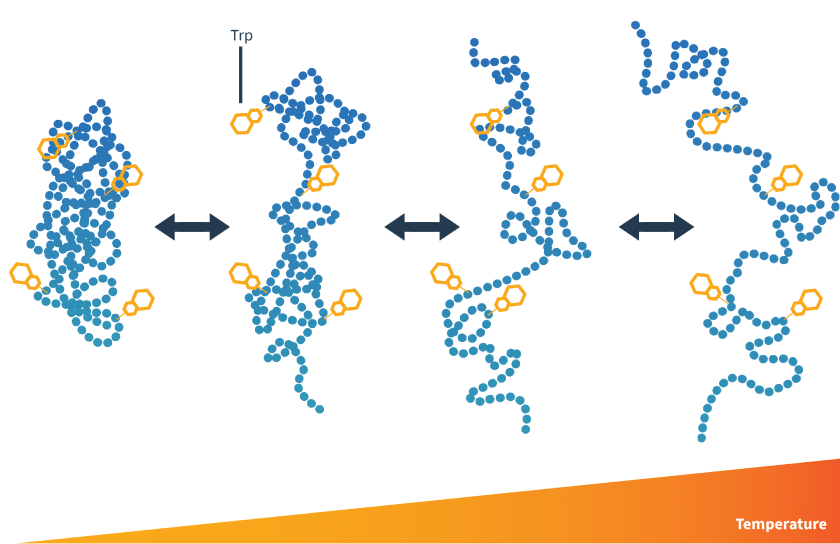
Tryptophan residues become exposed to solution in response to a temperature (or concentration) gradient

Proteins contain tryptophan residues, which absorb light at 280 nm. Their emission properties change depending on their chemical environment. When protected from aqueous solution, as when folded, they have a fluorescence emission peak at 330 nm.
When exposed to solution, the hydrophobic side chains have different emission properties, and have a fluorescence maximum at 350 nm. There is also an overall shift in tyrosine emission, which is useful in rare cases when a protein does not contain tryptophan residues.

First derivative overlaid onto the ratio vs gradient plot. The inflection point is easier to identify precisely by looking for the peak value.
Add nanoDSF to your workflow to learn more about your protein-based therapeutic samples

Prometheus offers several approaches to protein optimization that include nanoDSF optics. Learn more about the instrument and how it works.