Aggregation
Aggregation describes the formation of large clusters of biomolecules. Dianthus software helps to identify aggregates and particles since they are responsible for irregular TRIC-trace. The irregularity in TRIC-trace stems from larger particles traversing through the detection volume, thereby causing fluctuations in fluorescence intensity. In addition, larger aggregates typically sediment and accumulate at the bottom of the well, resulting in abnormal Well Scans. These wells are identified by the software and flagged as Well-Scan Quality.
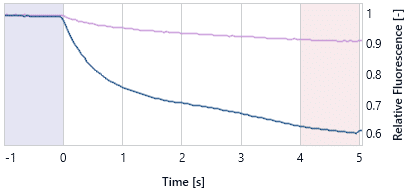
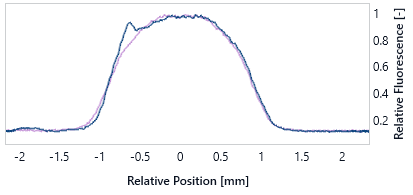
These figures show a typical TRIC trace (left) for an aggregated sample (dark blue) and a well scan for an aggregated sample (right) in comparison to a non-aggregate sample (purple). Note that Well Scan Anomalies can also be the result of air bubbles or bad mixing (see Liquid Handling).
Aggregated biomolecules often lose their functionality, leading to ambiguous results in many biochemical and biophysical assays. Aggregate detection by TRIC provides direct feedback on sample quality and thereby helps to establish optimal assay conditions. Aggregates interfere with data analysis and reduce reproducibility of TRIC experiments by increasing the level, and should be removed/prevented.
Aggregation of biomolecules can have different causes:
- Interaction of multimeric proteins: If multimeric proteins interact, the apparent formation of protein aggregates can be observed due to the formation of conglomerates. These multi-protein assemblies occur in a concentration dependent manner, and mostly at equimolar concentrations of both binding partners.
- Denatured or partially denatured molecules: If biomolecules such as proteins (partially) unfold, hydrophobic patches from the interior of the protein can become exposed to the surface. These hydrophobic patches can then interact with hydrophobic patches of other molecules to form aggregates. Partial unfolding can be caused by the presence of destabilizing agents, such as DMSO, ethanol, urea etc.
- Buffer pH: The pH of the buffer is critical for colloidal stability of proteins, since it directly affects the charge and conformational stability of the protein. Biomolecules usually have an overall positive or negative charge. If the buffer pH is equal to the Isoelectric Point of the molecule its net-charge is zero, which leads to a reduction of solubility in water. This allows the molecules to interact with each other rather than with surrounding water molecules, potentially resulting in the formation of aggregates. In general, pH values in the range of the isoelectric point (pI) of the protein should be avoided.
- Lack of stabilizing agents: Biomolecules are most stable in the presence of molecular crowding agents which mimic the physiological environment. For biochemical experiments, additives such as Detergents, polymers or amino acids are used to prevent aggregation by weakly interacting with the biomolecule surface.
- Lack of co-factors: Some biomolecules need co-factors (e.g. magnesium, calcium, heme) to maintain structural integrity. If they are deprived of these co-factors aggregation can occur.
- Inter-molecular disulfide formation: If case proteins contain surface-exposed cysteine residues, inter-molecular disulfide formation can result in aggregation. In such cases the addition of Reducing Agents such as 1-5 mM GSH or DTT is suggested.
- Inherently aggregation-prone biomolecules: Some biomolecules, e.g. membrane proteins with large, surface-exposed hydrophobic regions or intrinsically unfolded proteins, have a tendency to form aggregates. Solubilizing detergents should be used to keep these molecules in solution.
Recommendations:
- Aggregates are often caused by unspecific biomolecule interactions. The standard procedure to remove aggregates is a centrifugation step (30 min at >20.000 xg) and the addition of detergents or stabilizing additives, preferably 0.005% Tween® 20, 0.1% Pluronic® F-127 or 0.1% PEG 8000.
- Aggregation can be caused by the formation of unspecific disulfide bonds. We recommend using 2-5 mM Glutathion or DTT as a reducing agent to prevent disulfide bond formation.
- Aggregation is most pronounced when the biomolecules have a net charge of zero. To prevent aggregation the buffer pH should differ from the isoelectric point of the protein by at least 0.3 units. Since the exact isoelectric point of a protein is difficult to determine, it is recommended to empirically test different pH values.
- Aggregation can also be caused by too low salt concentrations that can result in charge-charge interactions of biomolecules. Concentrations of 150 mM NaCl, KCl or equivalent salts are recommended.
- The presence of multivalent cations such as Mg2+ can strongly promote aggregation of negatively charged biomolecules, e.g. DNA, and should be reduced to a minimum in these cases.